Current Research

Neural Biorepository Core
The Glioblastoma and Brain Metastasis Research Lab oversees the Neural Biorepository Core, an affiliate of the Houston Methodist Biorepository Core. The goal is to provide neurosurgery patient-derived tissue samples to facilitate translational and basic neuroscience research across the Texas Medical Center. Since its inception in 2018, samples from over 100 patients are collected yearly. In addition, Dr. Rostomily oversees comprehensive retrospective and prospective clinical databases related to brain and spinal metastases and gliomas.
To learn more about our core, visit here
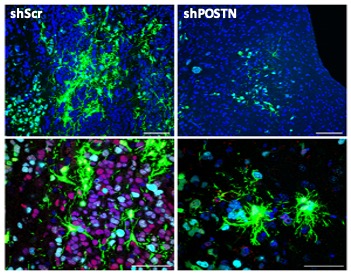
TWIST1 transcription factor function in glioblastoma (GBM)
TWIST1 is a master regulator of EMT that we first reported in GBM. Our subsequent studies demonstrated its functional importance for glioma malignancy and tumorigenicity in part through promoting stem cell and mesenchymal phenotypes. Loss of function studies support the potential therapeutic relevance of inhibiting TWIST1 or its regulated targets and pathways including Periostin (POSTN) and Akt among others. We also showed that site-specific TWIST1 phosphorylation and dimer motifs regulate gene expression and invasive phenotypes. Transcription factors present challenges to direct pharmacologic inhibition.
Therefore, in addition to basic mechanistic studies including novel protein-protein interactions, our current research is also addressing novel ways to target TWIST1 in part by leveraging these mechanisms of action. Current projects are focused on the roles of upstream regulators Dyrk1a and NR4A1/2 on TWIST1 function in GBM.
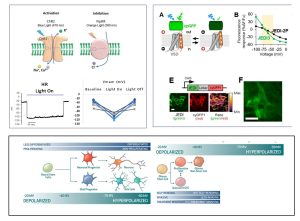
Electrophysiologic reprogramming of GBM cancer stem cells (GSCs)
In general, hyperpolarization is associated with differentiated and less proliferative cell states while depolarization is a property of more proliferative immature stem-like tissue and cancer cells. In collaboration with the St Pierre and Arenkiel labs at BCM and Clifford Stephan and Reid Powell at IBT (TAMU) we are pursuing Vmem modulation as a therapeutic target to “repgrogram” GSCs to less malignant and treatment responsive cell states. To establish the functional consequences of Vmem modulation we are employing 2 approaches- i) optogenetic regulation of Vmem in GSCs and ii) multiplexed high-throughput screens with Genetically Engineered Voltage Indicators (GEVIs) and cell cycle reporters (FUCCI and p27) to identify ion channel drugs that concurrently modify Vmem and cell cycle states.
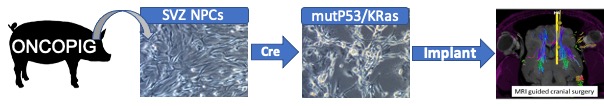
Large animal glioma model
In collaboration with Phil Horner and Amir Faraji we are developing a porcine syngeneic glioma model by orthotopic transplantation of transformed SVZ derived pig neural progenitors into SLA matched pigs. Unlike the rodent, the pig brain approximates the human in size and structure, thereby permitting more relevant studies of glioma growth patterns, imaging and surgical interventions. This project is further supported by collaborations with Larry Schook and Kyle Schachtschneider (Univ of Illinois) as well as HMRI CMP expertise and resources that permit clinical grade large animal surgery and MR imaging
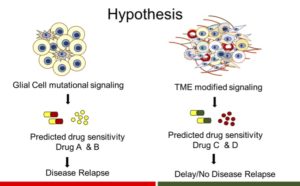
Intratumoral Molecular Heterogeneity
Intratumor molecular heterogeneity is a presumed result of selection pressure imposed by therapy and the tumor environment in the context of underlying tumor cell genomic instability. This heterogeneity has great implications for treatment resistance and tumor progression and the implementation of precision oncology. In collaboration with the Shendure Lab (University of Washington) we previously demonstrated intratumoral mutational heterogeneity in gliomas. Current studies involve the characterization and examination of the functional importance of heterogeneity of epigenetic modifications and matrix gene and protein expression in collaboration with Dr. Karol Bomsztyk (University of Washington)and Anil Korkut (MD Anderson), respectively.
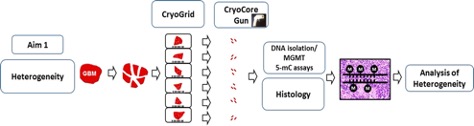
MGMT promoter methylation
Methylation of the MGMT promoter is a validated biomarker associated with longer survival after treatment with standard of care temozolomide and XRT “Stupp protocol”. However, the impact of different assays and pre-analytic tissue variables on the determination of methylation status have not been rigorously analyzed. In collaboration with Karol Bomsztyk (UW) and funded by an NIH U01 we are assessing the impact of different assays (methylation specific PCR, pyro-sequencing and MeDIP), fixation techniques, warm/cold ischemia time and intra-tumor heterogeneity on MGMT methylation in GBM.
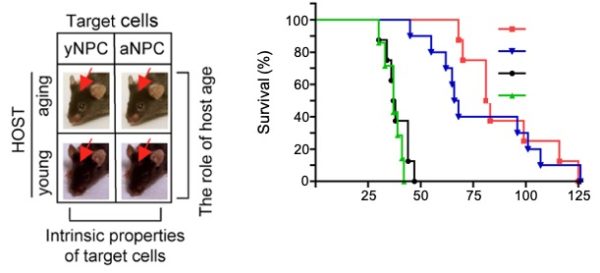
Aging and Glioma Malignancy
Aging is arguably the most robust prognostic factor for outcomes in human gliomas but the mechanisms of aging that contribute to glioma malignancy are not well understood. To address the lack of an aging appropriate glioma model to inform the study of aging mechanisms we generated a first in kind syngeneic glioma model employing transformed neural progenitor/stem cells from different aged donors implanted into different aged hosts. We observed age dependent cell intrinsic effects on survival associated with age dependent increased genomic instability, hypoxic tolerance and hypoxia regulated gene expression and resistance to TMZ and XRT which recapitulated key features of human age-dependent glioma phenotypes. Current research is aimed at further dissecting the roles of Hif1a and epigenetic changes as mechanisms regulating age-dependent genomic instability and glioma malignancy.
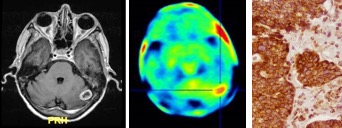
Brain Metastases
As their incidence increases and system therapies improve, brain metastases (BMs) are becoming a more prominent factor in outcome and quality of life for cancer patients. In collaboration with Drs. Steven Wong and Hong Zhao (co-PIs) we are studying mechanisms of astrocyte tumor cross-talk in brain metastasis as a potential therapeutic target. In addition, we are comparing expression and PET imaging of TSPO, a tumor and pro-inflammatory protein, as a potential biomarker of human brain metastasis malignancy and treatment responses.
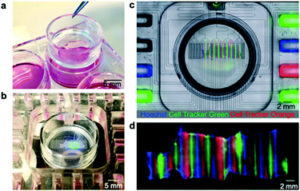
“Oncoslice”; Microfluidic Drug Delivery for Pre-Clinical Screening of Patient Derived Tumor Slice Cultures
The in silico predictions provided by genomic analyses do not always reliably predict actual patient treatment responses. In collaboration with the Folch lab and Ray Monnat at the University of Washington, Departments of Bioengineering and Genome Sciences we developed and tested a novel pre-clinical screening platform to bridge this gap. Using a microfluidic platform, multiple drugs are delivered to tumor derived slice cultures. Current research is focused on validation of predictive power in animal models, testing of immunotherapies and alternative tissue formats (3D bioprinting and tumor spheroids).
